apero_loc_spirou¶
Contents¶
1. Description¶
SHORTNAME: LOC
Localization calibration¶
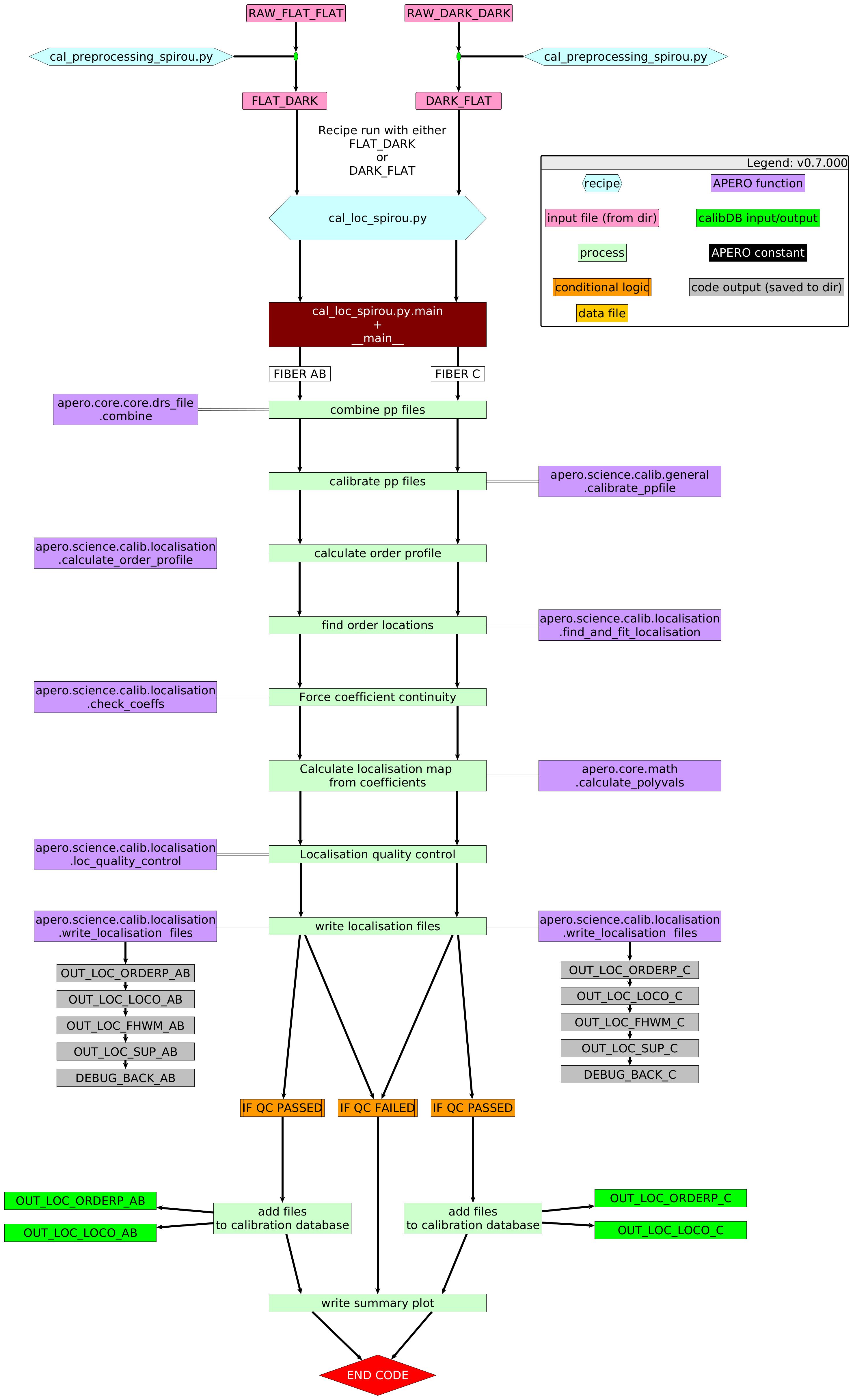
The localization recipe takes preprocessed DARK_FLAT or FLAT_DARK files (as many as given by the user or as many as occur on the nights being used via Aprocessing). It is run twice, once for the C fiber localization (with a set of DARK_FLAT) and once for the AB fiber localization (with a set of FLAT_DARK). It combines the DARK_FLAT files or the FLAT_DARK files into a single DARK_FLAT or FLAT_DARK (via a median combination of the images). After combining, the images are calibrated using our standard image calibration technique.
The first step in the localization code is to take the combined and calibrated DARK_FLAT or FLAT_DARK and apply a weighted box median, shown in equation:
where \(IM_{\text{orderp } j}\) is the order profile flux for all rows in the jth column, \(IM_{j=x:j=y}\) is the combined, calibrated DARK_FLAT or FLAT_DARK, that spans all columns from \(j=x\) to \(j=y\), and k is the column index number and ranges from \(j=0\) to \(j=4088\).
This produces the order profile image of the DARK_FLAT or FLAT_DARK which is used for the optimal extraction and to locate the orders.
To locate the orders we use the scikit measure.label algorithm which labels connected regions. Two pixels are defined as connected when both themselves and their neighbors have the same value. We use a connectivity value of 2 meaning that any of the 8 surrounding pixels can be neighbors if they share the same value.
In order to facilitate the labeling we first perform a 95th percentile of a box (of size \(25\times25\) pixels) across the whole image, as true illuminated pixels’ flux is location-dependent. We set a threshold at half that value and label all pixels above this threshold as one and all pixels below this to a value of zero. We then perform the measure.label on this Boolean map (referred to from this point on as \(Mask_{orders}\)). This is just a first guess of the order positions and usually returns many labeled regions that are not true orders.
To remove bad labels we first remove any labeled region with less than 500 pixels. We then remove any pixel within a labeled region that has a flux value less than 0.05 times the 95th percentile of all pixels in that given labeled region and remove this pixel from \(Mask_{orders}\). We then median filter each row of \(Mask_{orders}\) to clean up the labeled edges and apply a binary dilation (scipy ndimage.binary_dilation) algorithm. This binary dilation essentially merges labeled regions that are close to each other together by expanding regions marked with ones around the edges of these regions. After \(Mask_{orders}\) has been updated we re-run the labeling algorithm. As a final filtering step, we remove any region center that does not overlap with the central part of the image in the along-order direction (i.e., the center \(\pm\) half the width of the detector, 2044 \(\pm\) 1022 pixels).
Once we have the final set of labeled regions we use :math:`Mask_{orders} on each order to fit a polynomial fit (of degree 3) to the pixel positions in that labeled region forcing continuity between orders by fitting each coefficient across the orders. We also use the :math:`Mask_{orders} pixel positions to linearly fit the width of each order.
For a DARK_FLAT, this produces polynomial fits and coefficients for 49 orders for the C fiber. For a FLAT_DARK input, this produces polynomial fits and coefficients for 98 orders (49 orders for A and 49 orders for B). These polynomial coefficients for the positions of the orders and the widths of the orders are then converted into values as a function of position across each order.
As part of quality control we check that:
the number of orders is consistent with the required number of orders (49 for fiber C, 98 for fibers A+B).
the across-order value at the center of the detector is always larger than the value of the previous order
The order profile (ORDERP), locations of the orders (LOCO), and widths of the orders are saved to the calibration database (if both quality control criteria are met) for use throughout APERO.
2. Schematic¶
3. Usage¶
apero_loc_spirou.py {obs_dir}[STRING] [FILE:DARK_FLAT,FLAT_DARK] {options}
{obs_dir}[STRING] // [STRING] The directory to find the data files in. Most of the time this is organised by nightly observation directory
[FILE:DARK_FLAT,FLAT_DARK] // [STRING/STRINGS] A list of fits files to use separated by spaces. Current allowed types: DARK_FLAT OR FLAT_DARK but not both (exclusive)
4. Optional Arguments¶
--database[True/False] // [BOOLEAN] Whether to add outputs to calibration database
--badpixfile[FILE:BADPIX] // [STRING] Define a custom file to use for bad pixel correction. Checks for an absolute path and then checks 'directory'
--badcorr[True/False] // [BOOLEAN] Whether to correct for the bad pixel file
--backsub[True/False] // [BOOLEAN] Whether to do background subtraction
--combine[True/False] // [BOOLEAN] Whether to combine fits files in file list or to process them separately
--darkfile[FILE:DARKREF] // [STRING] The Dark file to use (CALIBDB=DARKM)
--darkcorr[True/False] // [BOOLEAN] Whether to correct for the dark file
--flipimage[None,x,y,both] // [BOOLEAN] Whether to flip fits image
--fluxunits[ADU/s,e-] // [STRING] Output units for flux
--plot[0>INT>4] // [INTEGER] Plot level. 0 = off, 1 = interactively, 2 = save to file
--resize[True/False] // [BOOLEAN] Whether to resize image
--no_in_qc // Disable checking the quality control of input files
5. Special Arguments¶
--xhelp[STRING] // Extended help menu (with all advanced arguments)
--debug[STRING] // Activates debug mode (Advanced mode [INTEGER] value must be an integer greater than 0, setting the debug level)
--listing[STRING] // Lists the night name directories in the input directory if used without a 'directory' argument or lists the files in the given 'directory' (if defined). Only lists up to 15 files/directories
--listingall[STRING] // Lists ALL the night name directories in the input directory if used without a 'directory' argument or lists the files in the given 'directory' (if defined)
--version[STRING] // Displays the current version of this recipe.
--info[STRING] // Displays the short version of the help menu
--program[STRING] // [STRING] The name of the program to display and use (mostly for logging purpose) log becomes date | {THIS STRING} | Message
--recipe_kind[STRING] // [STRING] The recipe kind for this recipe run (normally only used in apero_processing.py)
--parallel[STRING] // [BOOL] If True this is a run in parellel - disable some features (normally only used in apero_processing.py)
--shortname[STRING] // [STRING] Set a shortname for a recipe to distinguish it from other runs - this is mainly for use with apero processing but will appear in the log database
--idebug[STRING] // [BOOLEAN] If True always returns to ipython (or python) at end (via ipdb or pdb)
--ref[STRING] // If set then recipe is a reference recipe (e.g. reference recipes write to calibration database as reference calibrations)
--crunfile[STRING] // Set a run file to override default arguments
--quiet[STRING] // Run recipe without start up text
--nosave // Do not save any outputs (debug/information run). Note some recipes require other recipesto be run. Only use --nosave after previous recipe runs have been run successfully at least once.
--force_indir[STRING] // [STRING] Force the default input directory (Normally set by recipe)
--force_outdir[STRING] // [STRING] Force the default output directory (Normally set by recipe)
6. Output directory¶
DRS_DATA_REDUC // Default: "red" directory
7. Output files¶
name |
description |
HDR[DRSOUTID] |
file type |
suffix |
fibers |
dbname |
dbkey |
input file |
|---|---|---|---|---|---|---|---|---|
LOC_ORDERP |
Localisation: Order profile calibration file |
LOC_ORDERP |
.fits |
_order_profile |
AB, C |
calibration |
ORDER_PROFILE |
FLAT_DARK, DARK_FLAT |
LOC_LOCO |
Localisation: Position polynomial calibration file |
LOC_LOCO |
.fits |
_loco |
AB, C |
calibration |
LOC |
FLAT_DARK, DARK_FLAT |
LOC_FWHM |
Localisation: Width polynomial calibration file |
LOC_FWHM |
.fits |
_fwhm-order |
AB, C |
– |
– |
FLAT_DARK, DARK_FLAT |
LOC_SUP |
Localisation: Position superpositionimage calibration file |
LOC_SUP |
.fits |
_with-order |
AB, C |
– |
– |
FLAT_DARK, DARK_FLAT |
DEBUG_BACK |
Individual file background map |
DEBUG_BACK |
.fits |
_background.fits |
– |
– |
– |
DRS_PP |
8. Debug plots¶
LOC_WIDTH_REGIONS
LOC_FIBER_DOUBLET_PARITY
LOC_GAP_ORDERS
LOC_IMAGE_FIT
LOC_IM_CORNER
LOC_IM_REGIONS
9. Summary plots¶
SUM_LOC_IM_FIT
SUM_LOC_IM_CORNER
