science_seq¶
Contents¶
1. Description¶
Run all nightly science target steps.
This should be run after preprocessing of (at least) all science stars hot star files for all nights being reduced.
Include the following recipes:
EXTOBJ: Extracts all science target files
FTFIT1: Correct for telluric absorption in all science target files (without template)
FTTEMP1: Make a template for all science targets (using science files from FTFIT1)
FTFIT2: Correct for telluric absorption in all science target files (with template)
FTTEMP2: Make a template for all science targets (using science files from FTFIT2)
CCF: Calculate an estimate of RV based on the CCF for all science target files
POLAR: Calculate the polarization for all science target files (only calculated for those in polarimetry mode)
SCIPOST: Post-process all science target files
2. Schematic¶
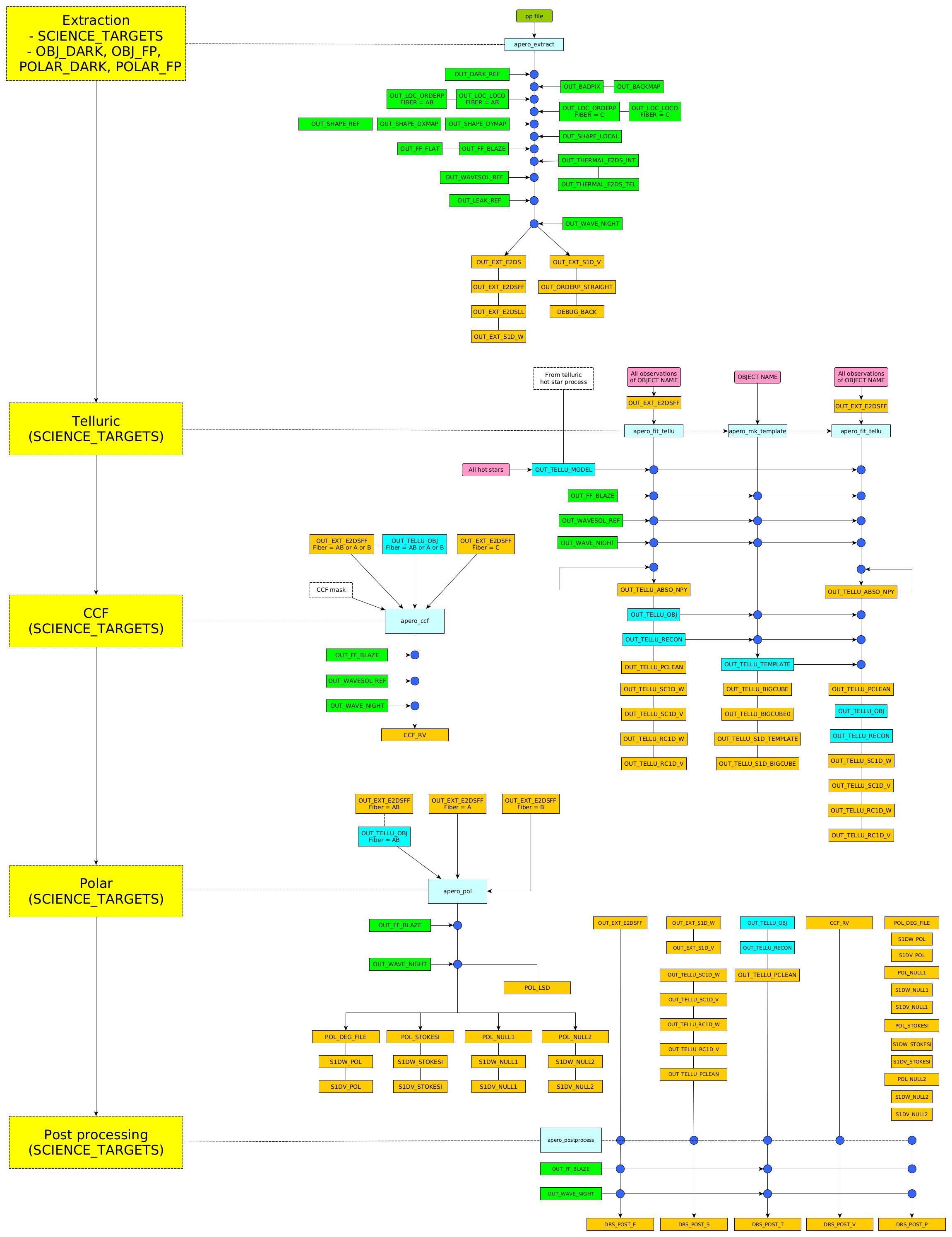
3. Recipes in sequence¶
ORDER |
RECIPE |
SHORTNAME |
RECIPE KIND |
REF RECIPE |
FIBER |
FILTERS |
ARGS |
KWARGS |
|---|---|---|---|---|---|---|---|---|
1 |
apero_extract_spirou.py |
EXTOBJ |
extract-science |
No |
– |
KW_OBJNAME: SCIENCE_TARGETS |
{files}=[OBJ_DARK, OBJ_FP, POLAR_DARK, POLAR_FP] |
|
2 |
apero_fit_tellu_spirou.py |
FTFIT1 |
tellu-science |
No |
AB |
KW_OBJNAME: SCIENCE_TARGETS |
{files}=[EXT_E2DS_FF] |
|
3 |
apero_mk_template_spirou.py |
FTTEMP1 |
tellu-science |
No |
AB |
KW_OBJNAME: SCIENCE_TARGETS |
||
4 |
apero_fit_tellu_spirou.py |
FTFIT2 |
tellu-science |
No |
AB |
KW_OBJNAME: SCIENCE_TARGETS |
{files}=[EXT_E2DS_FF] |
|
5 |
apero_mk_template_spirou.py |
FTTEMP2 |
tellu-science |
No |
AB |
KW_OBJNAME: SCIENCE_TARGETS |
||
6 |
apero_ccf_spirou.py |
CCF |
rv-tcorr |
No |
AB |
KW_DPRTYPE: OBJ_DARK, OBJ_FP, POLAR_DARK, POLAR_FP |
{files}=[TELLU_OBJ] |
|
7 |
apero_pol_spirou.py |
POLAR |
polar-tcorr |
No |
AB |
KW_DPRTYPE: POLAR_FP, POLAR_DARK |
–exposures=[TELLU_OBJ] |
|
8 |
apero_postprocess_spirou.py |
SCIPOST |
post-science |
No |
– |
KW_DPRTYPE: OBJ_DARK, OBJ_FP, POLAR_DARK, POLAR_FP |
{files}=[DRS_PP] |
