apero_wave_ref_spirou¶
Contents¶
1. Description¶
SHORTNAME: WAVEREF
Wavelength solution reference calibration¶
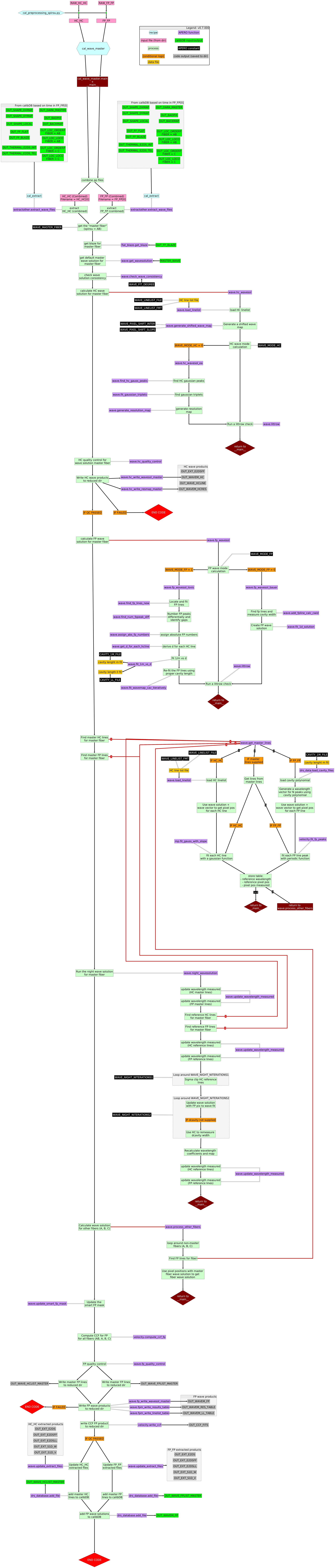
The wavelength solution generation follows the general idea of (Hobson et al 2021) however since publication there has been an overall reshuffling of the logic. As such we present an overview of the process here but refer the reader to (Hobson et al 2021) for further specific details.
The reference wavelength solution recipe takes preprocessed FP_FP and HC_HC files (as many as given by the user or as many as occur on the nights being used via apero_processing) from the reference night. It combines the FP_FP and HC_HC files into a single FP_FP and a single HC_HC file (via a median combination of the images). These combined FP_FP and HC_HC files are then extracted.
We first consider the combined flux in fibers A and B (the AB fiber). We locate the HC_HC lines, starting with a line list generated as in (Hobson et al 2021), fitting each peak with a Gaussian and measuring the position of the peak, and inferring peak wavelength from an initial guess at the wavelength solution from physical models. The first time this HC finding is performed we allow for a global offset between the current HC_HC file and the initial guess at the wavelength solution (this is important when our reference night is far in time from when our initial wavelength solution data was taken).
For the FP_FP AB fiber, a similar process is followed. However, instead of a single Gaussian, an Airy function is used (to account for the previous and following FP peak in the fitting process):
where F is the modeled flux of the FP, A is the amplitude of the FP peak, \(x_0\) is the central position of the FP peak, w is the period of the FP in pixel space, \(\beta\) is the shape factor of the FP peak and DC is a constant offset. Once we have found all HC and FP lines in the AB fiber we calculate the wavelength solution.
The accurate wavelength solution for reference night is then found through the following steps:
From FP peak spacing within each order, derive an effective cavity length per order.
Fit the chromatic dependency of the cavity with a 5th order polynomial and keep that cavity in a reference file; through the life of the instrument, we will assume that cavity changes are achromatic relative to this polynomial.
From the chromatic cavity solution, we find the FP order value of each peak, typically numbering from ~600 to ~24500 respectively at long and short wavelength ends of the SPIRou domain.
From the peak numbering, which is known to be an integer, we can refine the wavelength solution within each order. This solution is kept as a reference wavelength solution.
The finding of the fiber AB HC and FP lines and the calculation of the wavelength solution is repeated multiple times (in an iterative process). We essentially forget the locations of the HC and FP lines and re-find them as if we hadn’t found them before, only this time instead of the initial guess wavelength solution we use the previous iteration’s calculated solution and the previous iterations calculated cavity width fit as a starting point.
Finally, after three iterations, which is sufficient to converge to floating point accuracy, we re-find the HC and FP lines for the AB fiber one last time using the final reference wavelength solution and final cavity width fit. We also make an estimate of the resolution, splitting the detector into a grid of 3$times$3 and using all HC lines in each sector to estimate the line profile and thus the resolution of each sector. We then process each fiber (A, B, and C) in a similar manner to the AB fiber (finding HC and FP lines from the extracted images and calculating the wavelength solution) the only difference being we do not fit the cavity width nor do we fit the chromatic term; we force the coefficients to be the ones found with the AB fiber.
For quality control purposes we calculate an FP binary mask using the cavity width fit and use this to perform a cross-correlation function between the mask and the extracted FP for all fibers (AB, A, B, and C). We use the cross-correlation function to measure the shift of the wavelength solutions measured in fiber AB compared to fibers A, B, and C and confirm that this is less than 2 \(ms^{-1}\). As a second quality control, we match FP lines (found previously) between the fibers and directly calculate the difference in velocity between these lines as a second metric on the radial velocity shift between the fibers’ wavelength solutions. Note that typically for the reference night the value of these quality control metrics is around 10-20 \(cms^{-1}\) between fibers (i.e. \(AB-A\), \(AB-B\), \(AB-C\)).
The reference wavelength solution file (REFWAVE) for each fiber, a cavity fit file, and a table of all HC and FP lines found are then saved to the calibration database for use throughout APERO. A resolution map is also saved. The HC_HC and FP_FP extracted files have their headers updated with the reference wavelength solution.
2. Schematic¶
3. Usage¶
apero_wave_ref_spirou.py {obs_dir}[STRING] --hcfiles[FILE:HCONE_HCONE] --fpfiles[FILE:FP_FP] {options}
{obs_dir}[STRING] // [STRING] The directory to find the data files in. Most of the time this is organised by nightly observation directory
--hcfiles[FILE:HCONE_HCONE] // Current allowed types: HC1_HC1
--fpfiles[FILE:FP_FP] // Current allowed types: FP_FP
4. Optional Arguments¶
--database[True/False] // [BOOLEAN] Whether to add outputs to calibration database
--badpixfile[FILE:BADPIX] // [STRING] Define a custom file to use for bad pixel correction. Checks for an absolute path and then checks 'directory'
--badcorr[True/False] // [BOOLEAN] Whether to correct for the bad pixel file
--backsub[True/False] // [BOOLEAN] Whether to do background subtraction
--blazefile[FILE:FF_BLAZE] // [STRING] Define a custom file to use for blaze correction. If unset uses closest file from calibDB. Checks for an absolute path and then checks 'directory' (CALIBDB=BADPIX)
--combine[True/False] // [BOOLEAN] Whether to combine fits files in file list or to process them separately
--darkfile[FILE:DARKREF] // [STRING] The Dark file to use (CALIBDB=DARKM)
--darkcorr[True/False] // [BOOLEAN] Whether to correct for the dark file
--flipimage[None,x,y,both] // [BOOLEAN] Whether to flip fits image
--fluxunits[ADU/s,e-] // [STRING] Output units for flux
--locofile[FILE:LOC_LOCO] // [STRING] Sets the LOCO file used to get the coefficients (CALIBDB=LOC_{fiber})
--orderpfile[FILE:LOC_ORDERP] // [STRING] Sets the Order Profile file used to get the coefficients (CALIBDB=ORDER_PROFILE_{fiber}
--plot[0>INT>4] // [INTEGER] Plot level. 0 = off, 1 = interactively, 2 = save to file
--resize[True/False] // [BOOLEAN] Whether to resize image
--shapex[FILE:SHAPE_X] // [STRING] Sets the SHAPE DXMAP file used to get the dx correction map (CALIBDB=SHAPEX)
--shapey[FILE:SHAPE_Y] // [STRING] Sets the SHAPE DYMAP file used to get the dy correction map (CALIBDB=SHAPEY)
--shapel[FILE:SHAPEL] // [STRING] Sets the SHAPE local file used to get the local transforms (CALIBDB = SHAPEL)
--wavefile[FILE:WAVESOL_REF,WAVE_NIGHT,WAVESOL_DEFAULT] // [STRING] Define a custom file to use for the wave solution. If unset uses closest file from header or calibDB (depending on setup). Checks for an absolute path and then checks 'directory'
--forceext[True/False] // WAVE_EXTRACT_HELP
--cavityfile[FILE:WAVEREF_CAV] // WAVEREF_CAVFILE_HELP
--no_in_qc // Disable checking the quality control of input files
5. Special Arguments¶
--xhelp[STRING] // Extended help menu (with all advanced arguments)
--debug[STRING] // Activates debug mode (Advanced mode [INTEGER] value must be an integer greater than 0, setting the debug level)
--listing[STRING] // Lists the night name directories in the input directory if used without a 'directory' argument or lists the files in the given 'directory' (if defined). Only lists up to 15 files/directories
--listingall[STRING] // Lists ALL the night name directories in the input directory if used without a 'directory' argument or lists the files in the given 'directory' (if defined)
--version[STRING] // Displays the current version of this recipe.
--info[STRING] // Displays the short version of the help menu
--program[STRING] // [STRING] The name of the program to display and use (mostly for logging purpose) log becomes date | {THIS STRING} | Message
--recipe_kind[STRING] // [STRING] The recipe kind for this recipe run (normally only used in apero_processing.py)
--parallel[STRING] // [BOOL] If True this is a run in parellel - disable some features (normally only used in apero_processing.py)
--shortname[STRING] // [STRING] Set a shortname for a recipe to distinguish it from other runs - this is mainly for use with apero processing but will appear in the log database
--idebug[STRING] // [BOOLEAN] If True always returns to ipython (or python) at end (via ipdb or pdb)
--ref[STRING] // If set then recipe is a reference recipe (e.g. reference recipes write to calibration database as reference calibrations)
--crunfile[STRING] // Set a run file to override default arguments
--quiet[STRING] // Run recipe without start up text
--nosave // Do not save any outputs (debug/information run). Note some recipes require other recipesto be run. Only use --nosave after previous recipe runs have been run successfully at least once.
--force_indir[STRING] // [STRING] Force the default input directory (Normally set by recipe)
--force_outdir[STRING] // [STRING] Force the default output directory (Normally set by recipe)
6. Output directory¶
DRS_DATA_REDUC // Default: "red" directory
7. Output files¶
name |
description |
HDR[DRSOUTID] |
file type |
suffix |
fibers |
dbname |
dbkey |
input file |
|---|---|---|---|---|---|---|---|---|
EXT_E2DS_FF |
Extracted + flat-fielded 2D spectrum |
EXT_E2DS_FF |
.fits |
_e2dsff |
AB, A, B, C |
– |
– |
DRS_PP |
WAVESOL_REF |
Reference wavelength solution calibration file |
WAVESOL_REF |
.fits |
_wavesol_ref |
AB, A, B, C |
calibration |
WAVESOL_REF |
EXT_E2DS, EXT_E2DS_FF |
WAVEREF_CAV |
Reference wavelength cavity width polynomial calibration file |
WAVEREF_CAV |
.fits |
_waveref_cav_ |
AB |
calibration |
WAVECAV |
EXT_E2DS, EXT_E2DS_FF |
WAVE_HCLIST_REF |
Reference list of Hollow cathode lines calibration file |
WAVE_HCLIST_REF |
.fits |
_waveref_hclines |
AB, A, B, C |
calibration |
WAVEHCL |
EXT_E2DS, EXT_E2DS_FF |
WAVE_FPLIST_REF |
Reference list of FP liens calibration file |
WAVE_FPLIST_REF |
.fits |
_waveref_fplines |
AB, A, B, C |
calibration |
WAVEFPL |
EXT_E2DS, EXT_E2DS_FF |
WAVERES |
Reference wavelength resolution map file |
WAVE_RES |
.fits |
_waveref_resmap |
AB, A, B, C |
– |
– |
EXT_E2DS, EXT_E2DS_FF |
WAVEM_RES_E2DS |
Reference wavelength resolution e2ds file |
WAVEM_RES_E2DS |
.fits |
_waveref_res_e2ds |
AB, A, B, C |
calibration |
WAVR_E2DS |
EXT_E2DS, EXT_E2DS_FF |
CCF_RV |
Cross-correlation RV results file |
CCF_RV |
.fits |
_ccf |
AB, A, B, C |
– |
– |
EXT_E2DS_FF, TELLU_OBJ |
8. Debug plots¶
WAVE_WL_CAV
WAVE_FIBER_COMPARISON
WAVE_FIBER_COMP
WAVE_HC_DIFF_HIST
WAVEREF_EXPECTED
EXTRACT_S1D
EXTRACT_S1D_WEIGHT
WAVE_RESMAP
CCF_RV_FIT
CCF_RV_FIT_LOOP
9. Summary plots¶
SUM_WAVE_FIBER_COMP
SUM_CCF_RV_FIT
